MLR in R Exercise Solutions
- Fit the linear regression model
\[MORT=\beta_0+\beta_1*Lat+\beta_2*Long+\beta_{12}Lat*Long+\varepsilon_i\]
cancer.dat <- read.csv("Data/skincancer.csv",header=T)
cancer.lm <- lm(Mort ~ Lat*Long,data=cancer.dat)
summary(cancer.lm)##
## Call:
## lm(formula = Mort ~ Lat * Long, data = cancer.dat)
##
## Residuals:
## Min 1Q Median 3Q Max
## -34.649 -13.091 0.052 12.465 41.004
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 489.37255 185.66083 2.636 0.0115 *
## Lat -8.08381 4.49542 -1.798 0.0789 .
## Long -1.10396 1.98941 -0.555 0.5817
## Lat:Long 0.02318 0.04794 0.483 0.6312
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 19.36 on 45 degrees of freedom
## Multiple R-squared: 0.6857, Adjusted R-squared: 0.6647
## F-statistic: 32.72 on 3 and 45 DF, p-value: 2.233e-11- Obtain the confidence intervals for the regression parameters (the \(\beta\)’s).
confint(cancer.lm)## 2.5 % 97.5 %
## (Intercept) 115.43244935 863.3126482
## Lat -17.13804469 0.9704284
## Long -5.11084292 2.9029310
## Lat:Long -0.07339017 0.1197413- Create a plot of the residuals vs the fitted values.
cancer.resid <- residuals(cancer.lm)
cancer.fitted <- fitted(cancer.lm)
plot(cancer.fitted,cancer.resid,xlab="Predicted Values",ylab="Residuals",pch=16)
abline(h=0)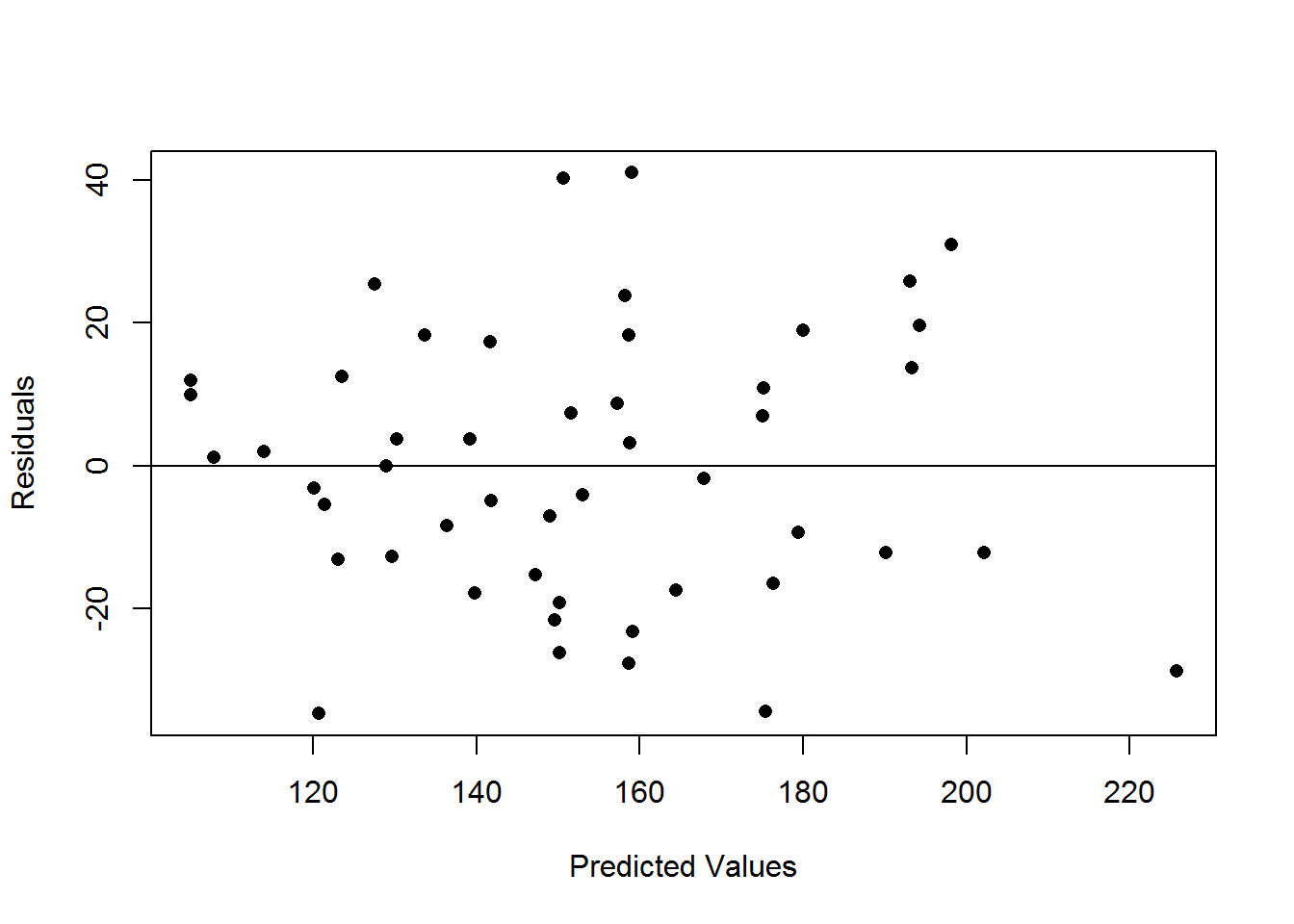 4. Create a histogram and QQ plot of the residuals.
4. Create a histogram and QQ plot of the residuals.
hist(cancer.resid,col="grey",main="Histogram of Residuals",freq=FALSE)
dens <- dnorm(seq(-40,45,by=0.2),mean=mean(cancer.resid),sd=sd(cancer.resid))
lines(seq(-40,45,by=.2),dens,col="red")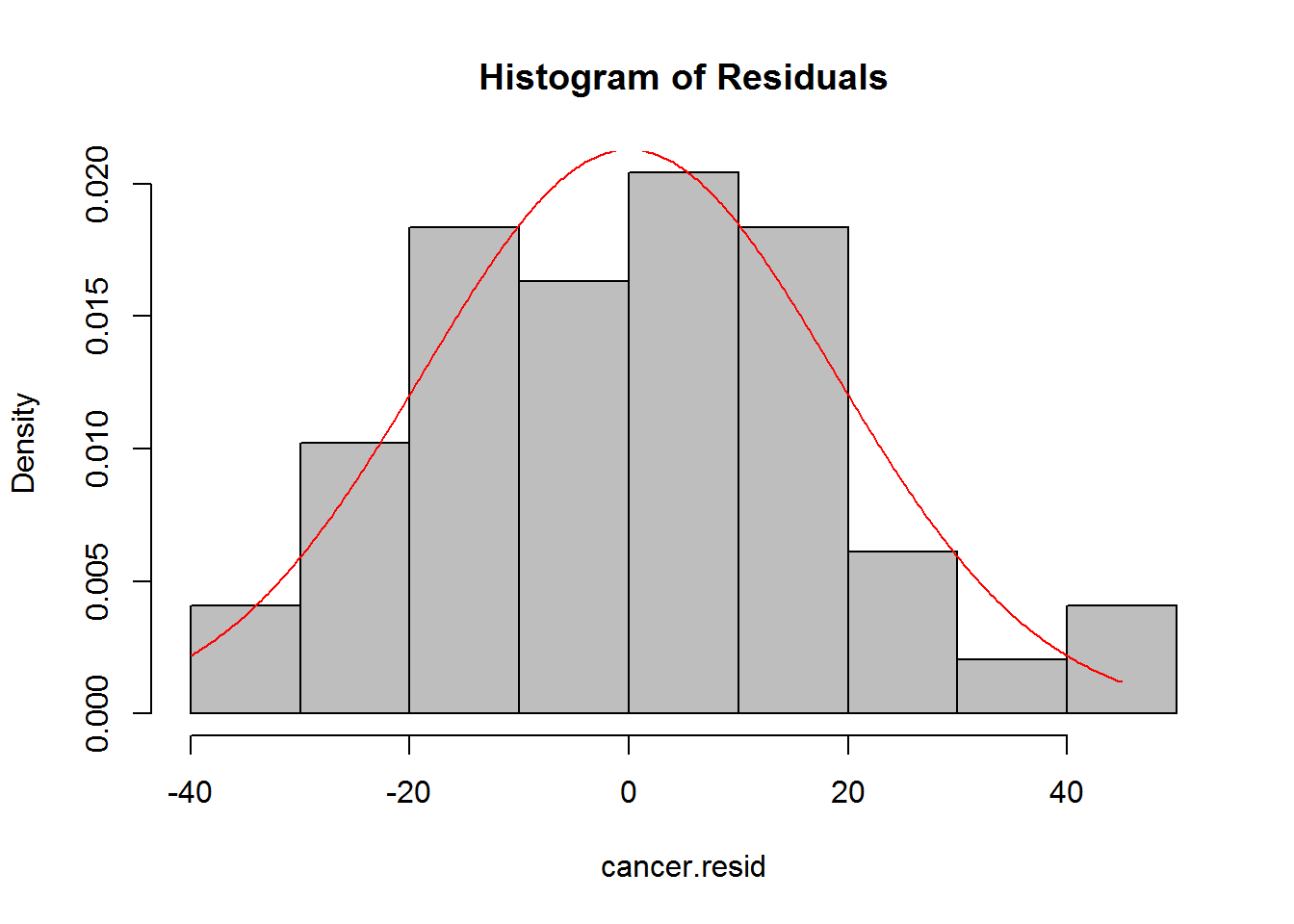
qqnorm(cancer.resid)
qqline(cancer.resid)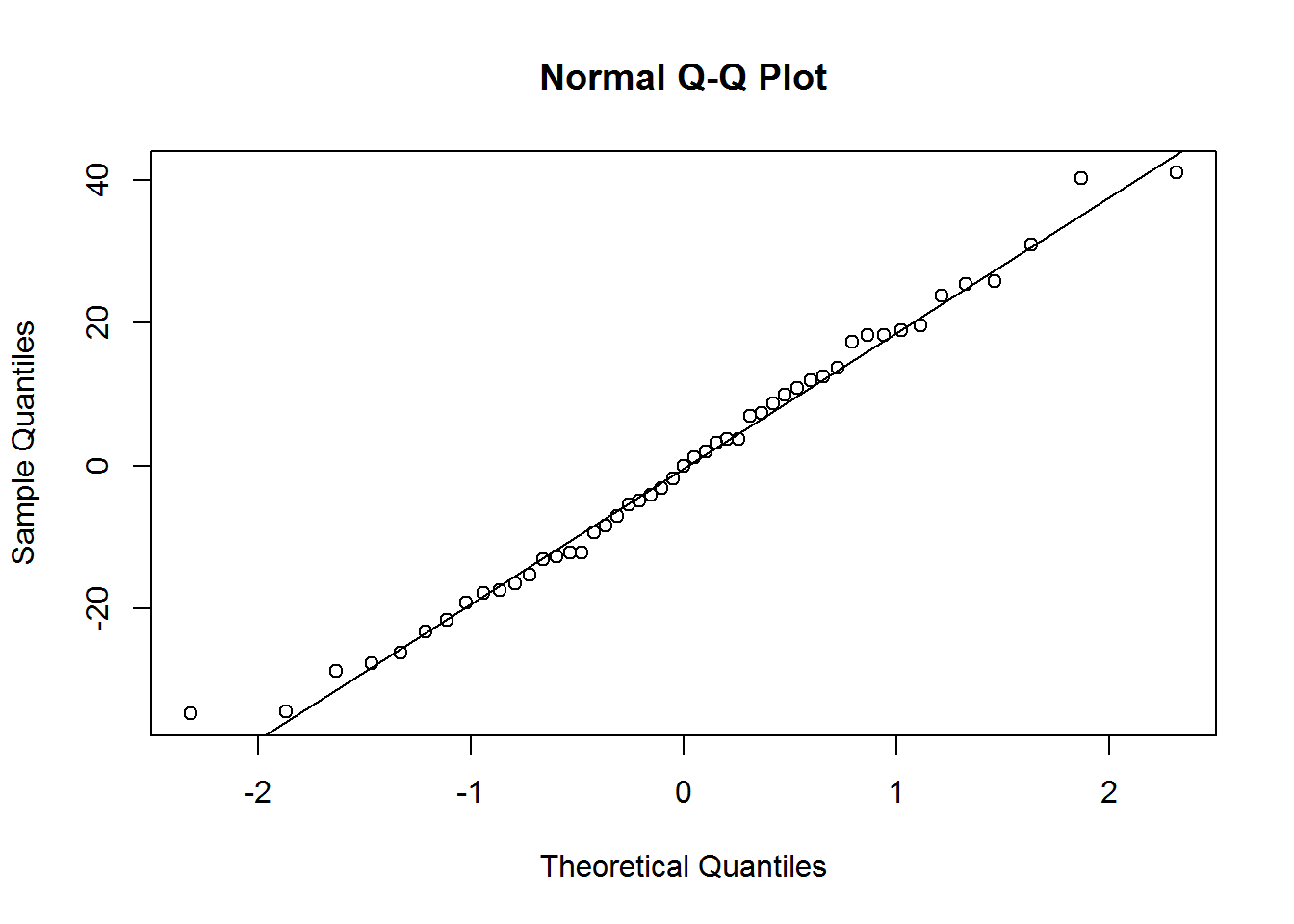 5. Calculate the confidence interval for the mean mortality rate and the prediction interval when Lat = 33 and Long = 86.
5. Calculate the confidence interval for the mean mortality rate and the prediction interval when Lat = 33 and Long = 86.
predict(cancer.lm,data.frame(Lat=33,Long=86),interval="confidence")## fit lwr upr
## 1 193.439 182.6452 204.2327predict(cancer.lm,data.frame(Lat=33,Long=86),interval="prediction")## fit lwr upr
## 1 193.439 152.9872 233.8907- Fit the reduced regression model without the interaction term.
cancer.lm2 <- lm(Mort ~ Lat+Long,data=cancer.dat)
summary(cancer.lm2)##
## Call:
## lm(formula = Mort ~ Lat + Long, data = cancer.dat)
##
## Residuals:
## Min 1Q Median 3Q Max
## -36.551 -13.525 -0.757 14.055 41.700
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 400.6755 28.0512 14.284 < 2e-16 ***
## Lat -5.9308 0.6038 -9.822 7.18e-13 ***
## Long -0.1467 0.1873 -0.783 0.438
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 19.19 on 46 degrees of freedom
## Multiple R-squared: 0.684, Adjusted R-squared: 0.6703
## F-statistic: 49.79 on 2 and 46 DF, p-value: 3.101e-12